At present there is a high demand of simple and reliable identification of the N501Y type coronavirus mutations which can be done in any lab.
Now, more than 1 year that the pandemic started and the many people infected, it is no surprise that covid mutants could be identified, given that this is in the nature of such viruses. This new coronavirus variants are at the suspicion to infect humans more effectively than the wild type and possibly may lead to more severe courses of the disease. Three major variants of SARS-CoV-2 are currently under the focus of attention, all belonging to the so-called N501Y mutation. These are the UK coronavirus variant N501Y.V1 (B 1.1.7 variant), the South African coronavirus N501Y.V2 and the Brazilian coronavirus N501Y.V3.
In order to keep infection rates low an accurate identification of the virus mutations in SARS-CoV-2 positive people is strongly needed. Point mutations are difficult to diagnose using qPCR tests. So far mutations are identified by genomesequencing of the virus. However, due to the sophisticated equipment and the long turn-around-time this is only done in very specialized laboratories from laboratory staff.
New Publication brings hope for fast and easy detection of Covid mutations
Just recently Kumar et al. published a paper on their development of a CRISPR based FnCas9 (Francisella novicida Cas9) method combined with the lateral flow dipstick Milenia HybriDetect called RAY (Rapid Variant AssaY). This test is designed for the simple and accurate detection of the British, South African and Brazilian coronavirus. Some months ago, in 2020, this group has also developed a test for the detection of the wild type of SARS-CoV-2 which is called FELUDA (FnCas9 Editor Linked Uniform Detection Assay). FELUDA is also a CRISPR based FnCas9 method and has been described in our blog earlier. The test developers already published in September 2020 that FnCas9 shows a very high specificity and could therefore be an ideal tool for the detection of point mutations. In their new paper they benefit from this feature of FnCas9 and were able to successfully detect all N501Y-type mutations with a simple paper strip test.
Development of RAY to detect coronavirus variant
Due to the lack of fast antigen based tests to identify SARS-CoV-2 mutant variants there is a high need of fast and cheaper mutant detection than the gold standard sequencing. The authors showed, that RAY can detect the covid infection and the presence of common N501Y mutation in one test with a high test sensitivity.
The researchers particularly concentrated on the SARS-CoV2 mutation N501Y, because it is present in all three new virus variants. A major step was to design a guide RNA (sgRNA) for the N501Y mutant, resulting in binding of FnCas9 to the mutant, but not to the wildtype.
First the group searched for the Cas9 PAM (Protospacer adjacent motif) sequence (NGG) in the surrounding area of the known mutations. The Cas9 protein requires the short specific PAM sequence to bind to the target sequence. The test developers recognized 10 sites targetable for FnCas9. One was the N501Y mutation which is present in all three new covid mutants.
Second they designed a FnCas9 sgRNA for the N501Y mutation. To design RAY, they took advantage of the fact, that FnCas9 is unable to bind targets with 2 mismatches at the PAM proximal 2nd and 6th position (which was previously reported). The wildtype of the virus is not addressed with this sgRNA and gives no signal.
The S-gene region addressed by the sgRNA for the detection of SARS-CoV2 used in the FELUDA test, which is not affected by the mutations, is used as an internal positive control.
Kumar et al. validated RAY on another SARS-CoV-2 mutation (T716I) and could show, that the principle of the test can be applied to other genetic variants of the virus.
SARS-CoV-2 Mutation Test on HybriDetect: RAY
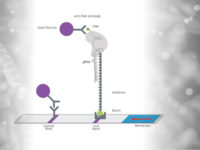
To improve the sensitivity and the test performance compared to electrophoresis based identification of the virus mutations, the test developers adapted RAY on Milenia HybriDetect lateral flow dipsticks (like FELUDA). Therefore, the reverse transcription and amplification of the S-gene region of SARS-CoV-2 containing N501Y mutation was done using biotinylated primers. The guideRNA was FAM-labeled (see Fig. x) and a catalytical inactive FnCas9 used. In a next step, the biotinylated amplicons and the RNP (gRNA +FnCas9) were incubated for 10 minutes at 37°C. To get a visible result on HybriDetect dipsticks, the sample was mixed with HybriDetect buffer and a dipstick placed for 2 to 5 minutes in the reaction.
For the covid mutation test, the FELUDA sgRNA is used in one reaction.
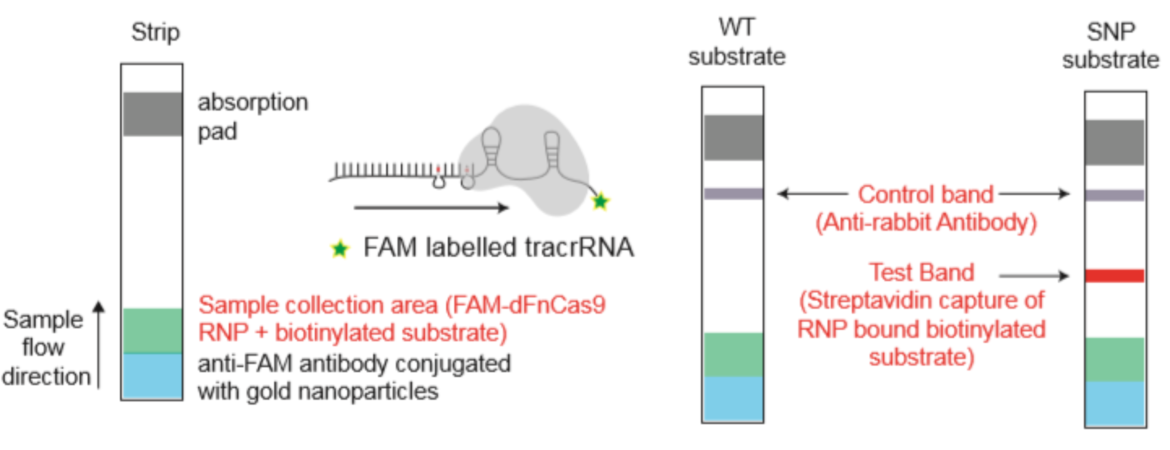
For the SARS-CoV-2 mutation test, the detecion by the S-gene sgRNA means, that the sample is SARS-CoV-2 positive. The detection by both sgRNAs – S-gene and N501Y – implicates, that a mutated variant of the coronavirus was detected in the sample.
The authors tested RAY on samples which were tested positive for SARS-CoV-2 by qPCR and sequenced for the detection of N501Y muation. The results showed, that RAY was able to identify all samples with genetic variants of the virus, even the samples with very high Ct values.
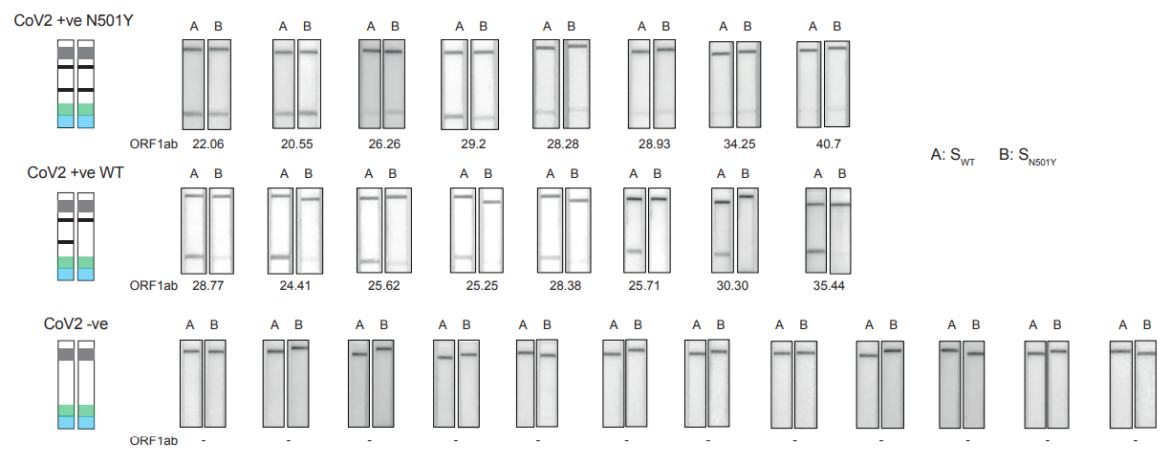
Advantages of RAY compared to sequencing tests
The test can be done with a regular thermocycler within 1 hour. The detection of the amplicons is done via our lateral flow strips and results can be reported within 5 minutes after starting the run of the test strips just by visual inspection. Therefore, the entire procedure from the time of taking the sample until the result can be reported takes only around 75 minutes. This test could be used for rapid diagnosis of mutated covid19 virus variants and therefore has the ability to increase test capacity which is highly needed. Still sequencing needs to be done to get information about new virus mutations occurring.
Do you want to know more about our products and news About Milenia Biotec? Please follow us on LinkedIn
https://www.linkedin.com/company/milenia-biotec

- Azhar, M., Phutela R., Kumar M., Ansari A.H., Rauthan R., Gulati S., Sharma N., Sinha D., Saumya S., Singh S., Acharya S., Paul D., Kathpalia P., Aich M., Sehgal P., Ranjan G., Bhoyar R., Singhal K., Lad H., Patra P.K., Makharia G., Chandak G.R., Pesala B., Chakraborty D., Maiti S: Rapid, accurate, nucleobase detection using FnCas9 https://doi.org/10.1101 (2020)
- Kumar M., Gulati S., Ansari A.H., Phutela R., Acharya S., Kathpalia P., Kanakan A., Maurya R., Vasudevan J.S., Murali A., Pandey R., Maiti S., Chakraborty D: RAY:CRISPR diagnostic for rapid and accurate detection pf SARS-CoV2 variants on a paper strip https://doi.org/10.1101/2021.02.01.21250900 (2021)