„CRISPR-Cas-based methods for simple, ultrasensitive and precise detection of nucleic acids have the potential to revolutionize the field of POC diagnostics.”
CRISPR-Cas & Lateral Flow
A Gene Editing Tool for Innovative Diagnostics

In 2020, Jennifer Doudna and Emmanuelle Charpentier received the Nobel Prize in chemistry for their scientific contributions to the simple and precise genome editing tool CRISPR-Cas. This has revolutionized biology within a decade as the most powerful tool for directed genetic manipulation. Together with the „CRISPR-pioneer“ Feng Zhang, the two women demonstrated the power of basic research. The molecular gene scissor is more topical than ever and promise great opportunities in areas such as medical therapy or plant breeding. Since then, the CRISPR-Cas toolbox has been continuously expanded and new application options presented. Therefore, it is not surprising that the CRISPR-Cas technology has found its way into molecular diagnostics. In combination with DNA amplification, this method offers impressive accuracy and great sensitivity, which is particularly attractive in the sector of Point-of-Care (POC) diagnostics.
History of CRISPR-Cas as Diagnostic Tools
In 2017 and 2018, Jonathan Gootenberg and Omar Abudayyeh (Broad Institute, Boston) showed that the gene editing tool “CRISPR-Cas” is also suitable for diagnostic purposes. They could demonstrate that the impressive specificity of the CRISPR system paired with the sensitivity of isothermal amplification techniques allows extremely precise and sensitive detection of nucleic acids. They called their innovative detection platform SHERLOCK, which stands for “Specific High-sensitivity Enzymatic Reporter unLOCKing”. In April 2018, two important publications showed that the SHERLOCK system can be combined with a simple, universal lateral flow device, the Milenia HybriDetect. This next logical step illustrates the potential of this technology as a simple, portable platform for applications outside of specialized laboratories. Almost at the same time, Janice Chen and colleagues from the working group around Nobel Prize winner Jennifer Doudna presented a SHERLOCK equivalent called DETECTR (“DNA-Endonuclease-Targeted CRISPR Trans Reporter”), which was based on the Cas12a protein and significantly simplified the overall reaction. The rapid pace of development of these methods was accelerated even more dramatically with the beginning of the pandemic. This short but impressive story is one reason why CRISPR-Cas-based methods have developed to one of the most exciting tools for POC-diagnostics.
Timeline: CRISPR-Cas as a diagnostic tool
Landmark Paper I
Gootenberg & Abudayyeh „Cas13a as diagnostic tool“ – SHERLOCK technology
Landmark Paper II
Chen et al. presenting DETECTR, simplified SHERLOCK equivalent using Cas12a
Landmark Paper III
Mhyrvold and Freije et al. presenting field-applicable SHERLOCK system using a HybriDetect
SARS-CoV-2 Pandemic
Start of the pandemic initiated massive boost of innovative molecular diagsnostics – Including CRISPR-Cas as diagnostic tools
More Techniques
methods using Cas9 and Lateral Flow described: CASLFA & FELUDA
Startups
Innovative Companies using CRISPR-Cas for diagnostics incresingly get into focus (Sherlock Biosciences, Mammoth Biosciences, CASPR Biotech)
1st FDA Approval
The Sherlock™ CRISPR SARS-CoV-2 kit receives FDA Emergency Use Authorization
Landmark Paper I
Gootenberg & Abudayyeh „Cas13a as diagnostic tool“ – SHERLOCK technology
Landmark Paper III
Mhyrvold and Freije et al. presenting field-applicable SHERLOCK system using a HybriDetect
More Techniques
methods using Cas9 and Lateral Flow described: CASLFA & FELUDA
1st FDA Approval
The Sherlock™ CRISPR SARS-CoV-2 kit receives FDA Emergency Use Authorization
Landmark Paper II
Chen et al. presenting DETECTR, simplified SHERLOCK equivalent using Cas12a
SARS-CoV-2 Pandemic
Start of the pandemic initiated massive boost of innovative molecular diagsnostics – Including CRISPR-Cas as diagnostic tools
Startups
Innovative Companies using CRISPR-Cas for diagnostics incresingly get into focus (Sherlock Biosciences, Mammoth Biosciences, CASPR Biotech)
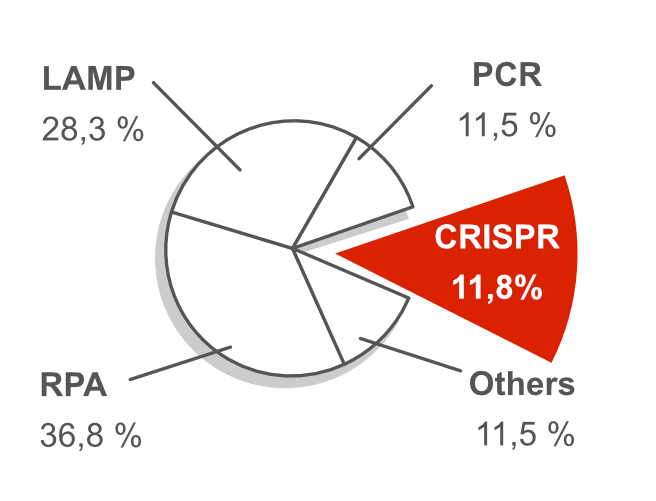 An important methodological trend of the last decade is the combination of DNA amplification and lateral flow based readout. The analysis of more than 300 scientific publications revealed that the CRISPR-Cas toolbox became one the most relevant topics for innovative, molecular POC-diagnostics.
An important methodological trend of the last decade is the combination of DNA amplification and lateral flow based readout. The analysis of more than 300 scientific publications revealed that the CRISPR-Cas toolbox became one the most relevant topics for innovative, molecular POC-diagnostics.
General Mechanism of CRISPR-Cas based Methods
An Expanding Toolbox and Two General Detection Strategies
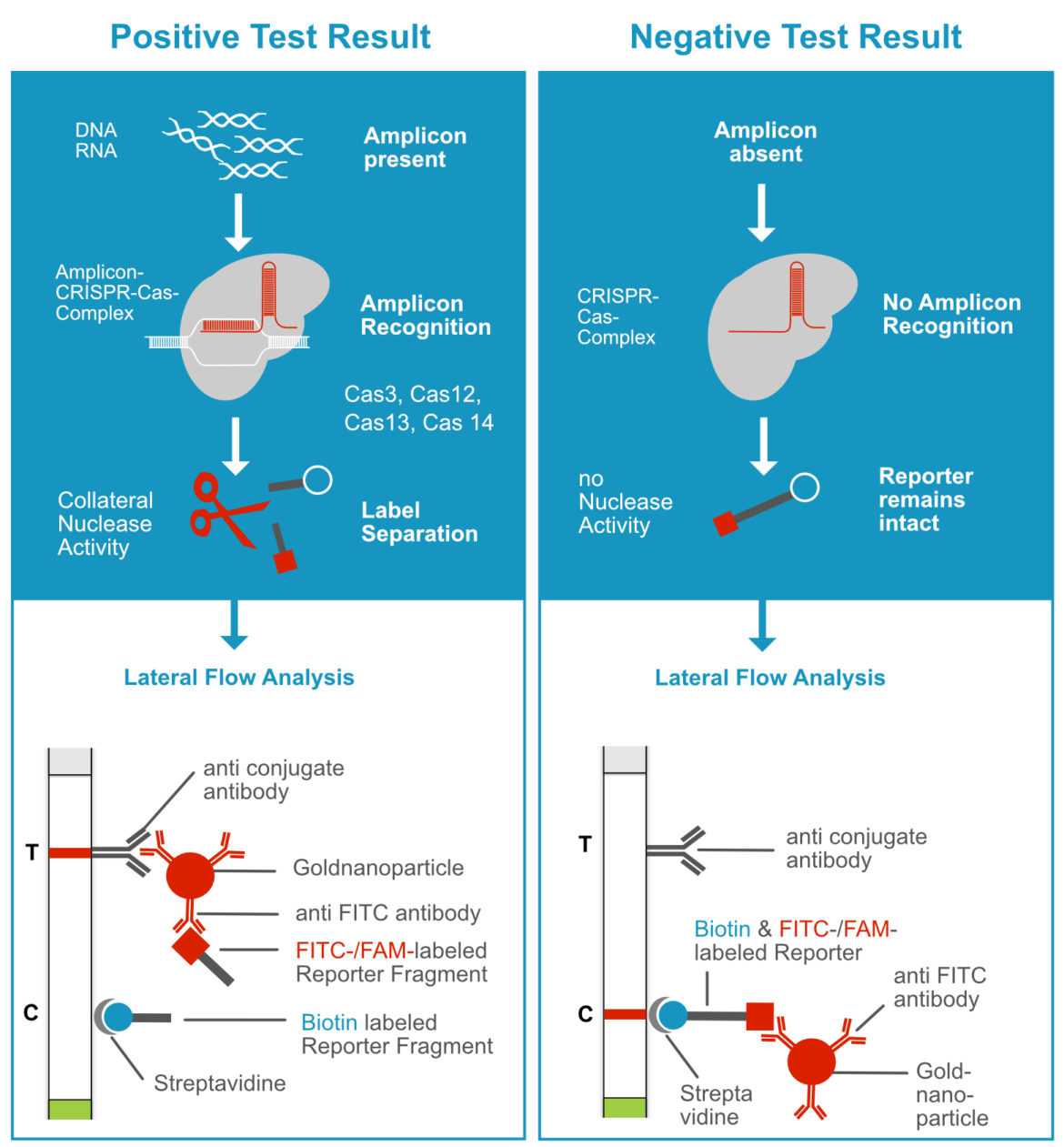
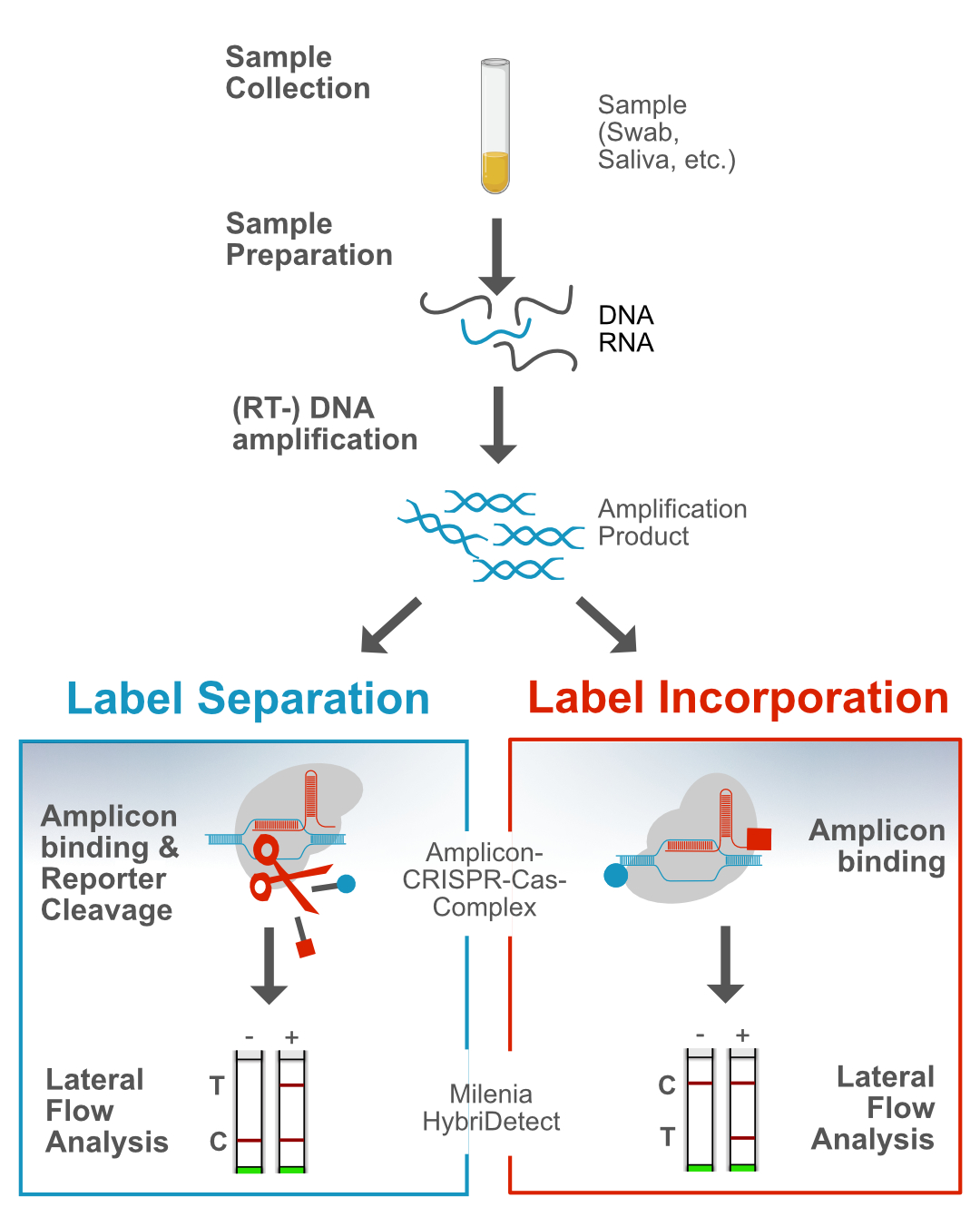 Interestingly, special CRISPR-associated proteins (Cas proteins) with diagnostically relevant characteristcs were used for methods like SHERLOCK or DETECTR. The CRISPR-Cas-complexes are able to “recognize” amplicons with an excellent specificity. This in turn leads to the activation of a collateral nucelase activity of the Cas-Protein. As a result, terminal labeled reporter molecules (Cas12a: ssDNA; Cas13a: ssRNA) are efficiently cleaved by the Cas-protein. The seperation of terminal labels is detectable with the Milenia HybriDetect. This mechanism can be understood as an additional enhancer reaction beside DNA amplification. Based on this, numerous methodological variants have been developed, but the general detection mechanism is ultimately based on the cleavage of a reporter. Therefore, we define this general detection mechanism as „Reporter Degradation Strategy“ or „Label Separation Strategy“.
Interestingly, special CRISPR-associated proteins (Cas proteins) with diagnostically relevant characteristcs were used for methods like SHERLOCK or DETECTR. The CRISPR-Cas-complexes are able to “recognize” amplicons with an excellent specificity. This in turn leads to the activation of a collateral nucelase activity of the Cas-Protein. As a result, terminal labeled reporter molecules (Cas12a: ssDNA; Cas13a: ssRNA) are efficiently cleaved by the Cas-protein. The seperation of terminal labels is detectable with the Milenia HybriDetect. This mechanism can be understood as an additional enhancer reaction beside DNA amplification. Based on this, numerous methodological variants have been developed, but the general detection mechanism is ultimately based on the cleavage of a reporter. Therefore, we define this general detection mechanism as „Reporter Degradation Strategy“ or „Label Separation Strategy“.
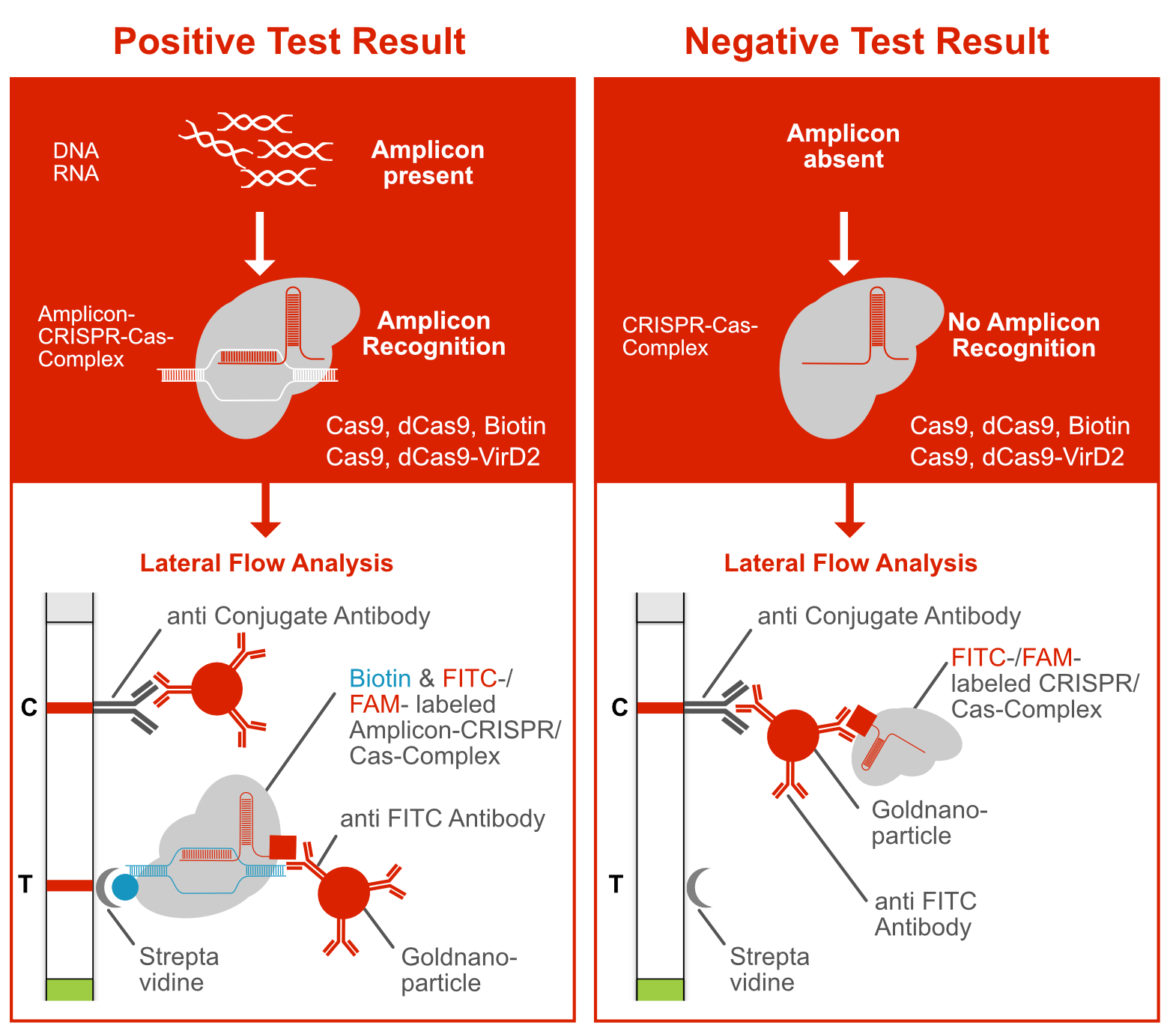
Since 2020, an increasing number of papers have been published that use the CRISPR-Cas-toolbox but do not follow the „Reporter Degradation Strategy“. The Cas9 protein, may be the best-known representative of the Cas protein family, moves into the focus of these methods. The complex of a chimeric guide RNA and Cas9 enables the specific “recognition” of the amplicon. This amplicon-CRISPR-Cas9 complex is detectable via Lateral Flow using the Milenia HybriDetect. Therefore, we define this methodological approach as “Label Incorporation Strategy“.
Both general detection strategies allow a simple, fast readout through lateral flow analysis. However, the use and interpretation of the universal Lateral Flow Device has to be adapted to the used method.
How to modify a CRISPR/Cas-based assay for Lateral Flow Readout
The adaptation of your own assay depends very much on the selected test method. If a detection system such as SHERLOCK or DETECTR is used in accordance with the „Label Separation Strategy“, the dual labeled reporter molecule is the most important component that needs to be questioned. The choice of the optimal reporter sequence depends on the CRISPR-Cas system used. Ultimately, it is important to determine the exact reporter concentration which works for the nuclease reaction and for the lateral flow analysis. The following table gives a brief overview of already described reporters for Cas13a and Cas12a dependent test formats.
| Cas Protein | Reporter Type | 5' Label | 3' Label | Sequence (5' > 3') |
|---|---|---|---|---|
| LwaCas13a | ssRNA | 6-FAM | Biotin | poly-U (≥ 10) |
| LwaCas13a | ssRNA | 6-FAM | Biotin | (RU)14 |
| LwaCas13a | ssRNA | 6-FAM | Biotin | (MARARUGRGRCMA)2 |
| LbCas12a | ssDNA | 6-FAM | Biotin | TTATT |
| LbCas12a | ssDNA | 6-FAM | Biotin | TTATTATT |
| LbCas12a | ssDNA | 6-FAM | Biotin | (TTATT)3 |
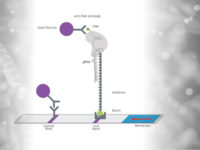
If the detection is based on the recognition of the amplicon-CRISPR-Cas-Complex, various labeling strategies can be followed. In this scenario, guide RNA, Cas protein and amplicon usually come together to form a detectable complex including the Labels biotin and FITC/FAM. The following figure describes three possible labeling strategies that have already been successfully published.
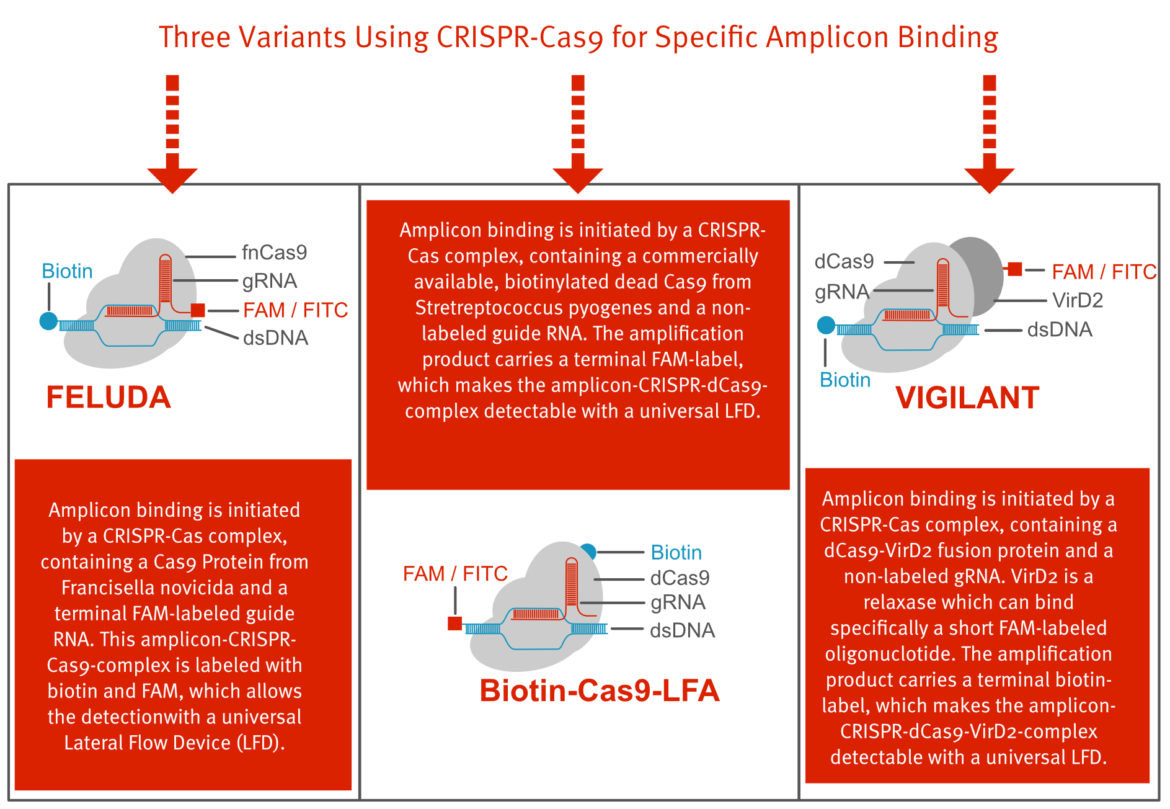
Considering the fact that CRISPR-Cas, as a diagnostic tool, was combined for the first time with a simple lateral flow readout in 2018, the actual methodological variety is absolutely impressive. Only 3 years later there are numerous modifications and the toolbox is constantly expanding. Some important techniques that were described in the past are listed in the table below.
| Method Name (Acronym) | General Detection Strategy | Amplification Method | Cas Protein (Origin) | Labeling Strategy | Lateral Flow Device | Reference |
|---|---|---|---|---|---|---|
| FELUDA | Amplicon Binding Assay | PCR | FnCas9 (Francisella novicida) | Amplicon (Biotin), gRNA (FAM) | Milenia HybriDetect | Azhar et al., 2020 |
| SHERLOCK | Reporter Degradation Assay | LAMP, RPA (and in vitro transcription) | e.g. LwaCas13a (Leptotrichia wadei) | Collateral Cleavage of FAM-Biotin-Reporter | Milenia HybriDetect | Kellner et al., 2020 |
| CASLFA | Amplicon Binding Assay | PCR, RPA | Cas9 (and dCas9, origin unkown) | Probe-functionalized Nanoparticles, Amplicon (Biotin) | Selfmade LFD | Wang et al., 2020 |
| DETECTR | Reporter Degradation Assay | LAMP, RPA | e.g. LbCas12a (Lachnospiraceae bacterium) | Collateral Cleavage of FAM-Biotin-Reporter | Milenia HybriDetect | Broughton et al., 2020 |
| VIGILANT | Amplicon Binding Assay | RPA | VirD2*-SpdCas9** fusion protein (*Agrobacterium tumefaciens, **Streptococcus pyogenes) | VirD2-dCas9 (FAM), Amplicon (Biotin) | Milenia HybriDetect | Marsic et al., 2021 |
| iSCAN | Reporter Degradation Assay | LAMP | LbCas12a (Lachnospiraceae bacterium) | Collateral Cleavage of FAM-Biotin-Reporter | Milenia HybriDetect | Ali et al., 2020 |
| "Biotin-dCas9-LFA" | Amplicon Binding Assay | RPA | SpdCas9* (Streptococcus pyogenes, *3xFLAG + Biotin) | SpdCas9 (Biotin), Amplicon (FITC) | Milenia HybriDetect | Osborn et al., 2021 |
Pros & Cons using CRISPR-Cas
Specificity / Selectivity
One of the most important advantages of CRISPR-Cas-based nucleic acid detection is the impressive selectivity. By preselecting a suitable gRNA / crRNA, it is possible to discriminate between sequences with only one differing base pair. This specificity down to single base level could be shown for the SHERLOCK (-like) applications as well as for CRISPR-Cas9-dependent amplicon binding assays.
Sensitivity
Especially Cas proteins with collateral nuclease activity (Cas12, Cas13, Cas14, Cas3, etc.) can significantly contribute to an increase in detection sensitivity. The combination of a very sensitive DNA amplification technique with a CRISPR-Cas-induced reporter degradation can be seen as two consecutive enhancer reactions. This fact allows the very precise detection of DNA or RNA, even at attomolar concentrations. Current publications indicate that Cas protein modification can further improve the test performance.
Assay-Complexity
Most CRISPR-Cas-based systems for the detection of nucleic acids are based on the combination of DNA amplification and amplicon recognition by the effector complex. This point can certainly be discussed critically. In order to develop a very sensitive test, it is essential to establish a pre-amplification reaction. Many POC-relevant questions can already be answered with “simple” DNA amplification methods combined with Lateral Flow. The use of CRISPR-Cas components can complicate assay development significantly. For this reason, it should be clearly defined whether the requirements for the test justify the use of this platform before the development starts.
Multiplexing
At this point, a clear distinction have to be made between the two CRISPR-dependent detection strategies described. „Progammable“ Cas proteins with collateral nuclease activity, such as Cas12 or Cas13, can significantly improve the sensitivity of a nucleic acid detection assay. Nevertheless, reporter degradation is indiscriminate, which excludes multiplexing using a single Cas protein. A certain degree of multiplexing can be achieved by using different Cas proteins with deviating „cutting“ preferences. However, this complicates the assay development considerably. In contrast, the use of the CRISPR-Cas9 amplicon binding concept certainly allows multiplexing. Especially, the VIGILANT method needs to be highlighted. Direct labeling of the Cas9-VirD2 fusion protein can massively reduce false positive results due to undesired interaction of labeled nucleic acids. Furthermore, this system is perfectly compatible with the Milenia HybriDetect 2T, a universal Lateral Flow Device allowing simultanious detection of 2 target.
| Method combined with LFA | Sensitivity | Specificity | Speed | reverse Transcription (one pot) | Robustness | Method. Variety | Simple Assay Design | Multiplexing Ability | Prevention Carryover Cont. |
|---|---|---|---|---|---|---|---|---|---|
| PCR | |||||||||
| LAMP | |||||||||
| RPA | |||||||||
| CRISPR (Label-Sep.) | |||||||||
| CRISPR (Label-Inc.) |